2023
Head direction (HD) cells are neurons that activate when an animal faces a specific direction in the horizontal plane. Together, these neurons form a neuronal compass, playing a central role in spatial representation. While originally discovered in rodents [1], HD cells have since been identified in various species, including flies [2] and more recently in zebrafish larvae [3]. HD circuits are thought to have a the architecture of a ring attractor, and thus to exhibit a 1D continuum of stable configurations, each corresponding to a distinct orientation [4].
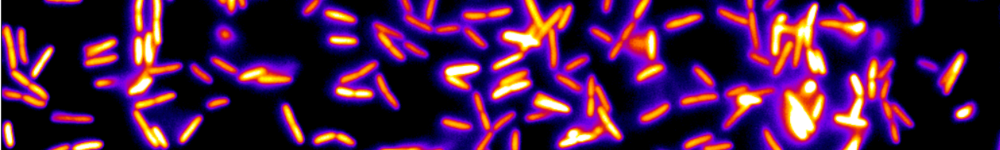
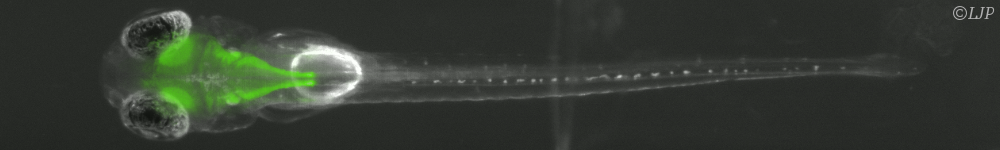
In this project, we aim to explore the fundamental principles underlying the organization and function of HD circuits using Danionella cerebrum (DC), a recent introduced model vertebrate system [5]. DC is a small freshwater fish whose brain remains small and almost entirely transparent until adulthood. This exceptional trait enables us to monitor the brain's complete activity with cellular resolution across development using calcium imaging.
During the internship, the candidate will use a virtual reality setup developed in our lab to record brain activity as the fish fictively navigates within a virtual environment (see figure 1). The intern will design and implement specific protocols and develop analysis methods to functionally identify HD cells. Once identified, he/she will study how the HD cell circuit matures during development to acquire the specific properties of a ring attractor. We further aim to reveal how multiple sensory inputs, including visual, vestibular, and proprioceptive cues, converge to drive the dynamics of the HD circuit, ensuring the accurate representation of the animal's orientation.
Context: the Laboratoire Jean Perrin (LJP) is a biophysics lab located on the P&M Curie campus. Our research group, which currently comprises 3 permanent researchers, 4 PhD students, and 1 post-doc, focuses on unraveling the neuronal basis of sensory-driven and spontaneous behavior using Zebrafish and Danionella as model vertebrates [6]. We develop custom bio-imaging setups for whole-brain functional recording and optogenetic activation while performing quantitative behavioral assays. Additionally, we dedicate substantial effort to the development of computational methods, inspired from statistical physics and machine-learning approaches, for modeling and analyzing these large datasets.
[1] J.S. Taube, R.U. Muller, J.B. Ranck Jr, J. Neurosci. 10(2):420-35 (1990)
[2] Seelig, J.D., and Jayaraman, Nature 521, 186–191 (2015)
[3] Petrucco et al., Nature neuroscience, volume 26, pages 765–773 (2023)
[4] Kim, S.S., Rouault, H., Druckmann, S., and Jayaraman, V. Science 356, 849–853 (2017)
[5] Schulze et al. Nature Methods volume 15, pages 977–983 (2018)
[6] https://www.labojeanperrin.fr/?article6