Morphogenesis
André Estevez-Torres, Jean-Christophe Galas, Baptiste Blanc, Nicolas Lobato-Dauzier, Romain Leroux
Former members: Guillaume Sarfati, Marc Van Der Hofstadt, Yuliia Vyborna, Anis Senoussi, Shunichi Kashida, Georg Urtel, Anton Zadorin, Jonathan Lee Tin Wah, Adrian Zambrano
News: We have just obtained an ANR funding (LiliMat - Life-Like Materials, 2022-2026) as well as a PEPR funding (MoleculArxiv - DNA synthesis for data storage). We had obtained in 2018 an ERC consolidator grant entitled 'Living-inspired metabolic soft matter'.
You may find further information in our detailed webpage: http://ljpmorpho.free.fr
We are engineering synthetic chemical systems that reproduce essential dynamic features of networks and populations in biology. The objective of our biomimetic approach is two-fold. On the one hand, by studying simple, physics-like and controllable molecular systems that retain the essential features of their biological analogues we hope to provide new insights on the emergence of complex biological behaviors -such as gene regulation, and morphogenesis. On the other hand, these dynamic molecular systems can be regarded as a new kind of "life-like" materials capable of adapting and responding autonomously to their environment.
To this end we essentially use systems based on nucleic acid hybridization reactions because their reactivity can be easily predicted by, roughly, Watson-Crick pairing rules.
More precisely we work with four (mainly 1 and 2) powerful and complementary experimental systems.
- 1) The PEN DNA toolbox, a molecular programming language based on short DNA strands and three enzymes that can be kept out of equilibrium for tens of hours (see figure 1 below).
- 2) The microtubule/kinesin system, a versatile active matter system that performs chemo-mechanical transductions (see figure 2 below).
- 3) Cell-free transcription translation systems to study in vitro de dynamics of gene regulatory networks.
- 4) DNA self-assembled nanostructures called DNA origami.
We further develop microfluidic and micropatterning techniques for the spatial and temporal control of these reactive systems.
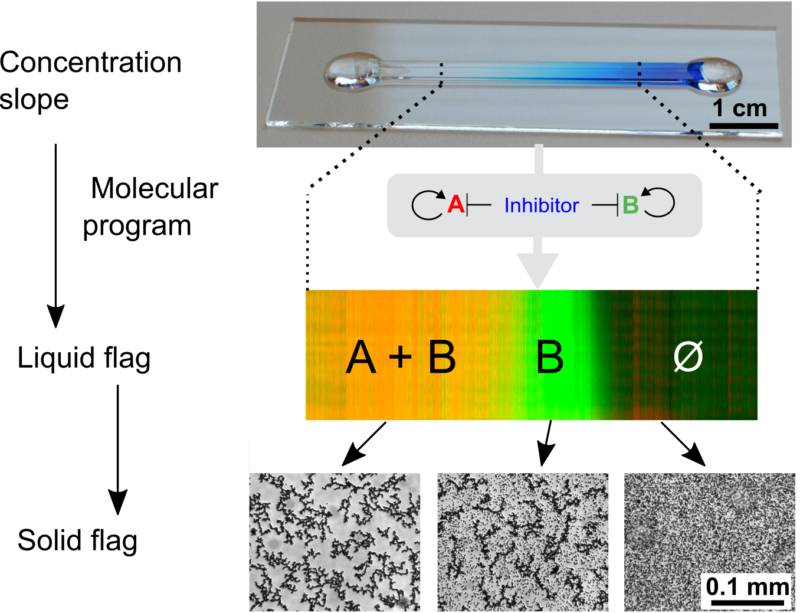
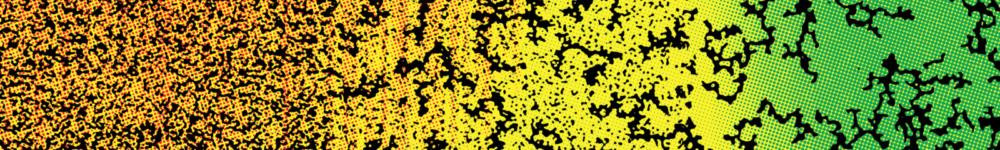
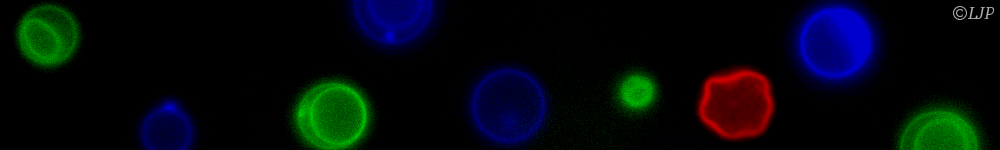
Figure 1: A "life-like" material through reaction-diffusion patterning. A molecular program based on short DNA strands and enzymes is able to read and interpret a pre-existing gradient of immobile DNA to generate several chemically distinct zones. As for cells in an embryo, the fate of DNA-coated beads (translated into a level of aggregation) depends on the position.
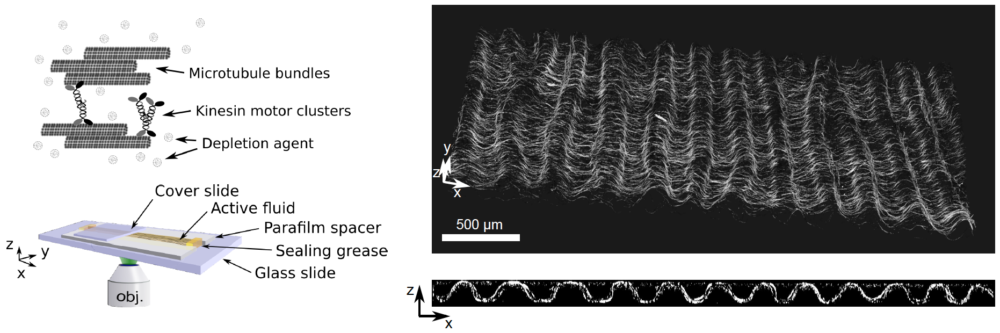
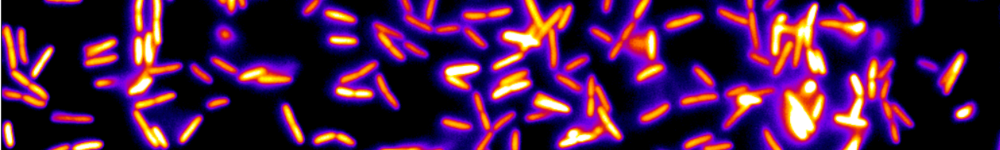
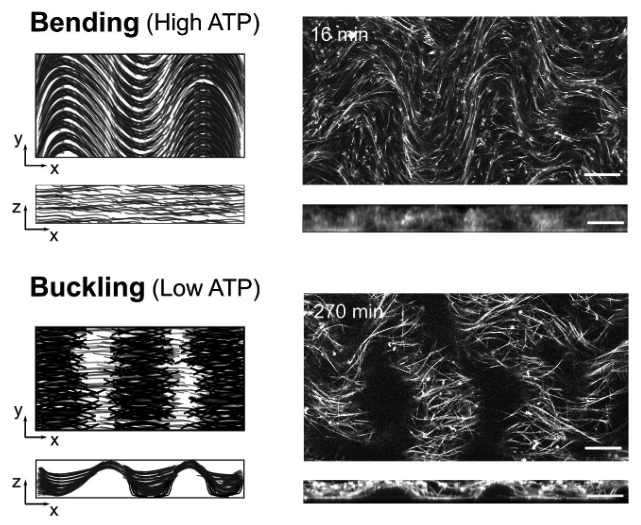
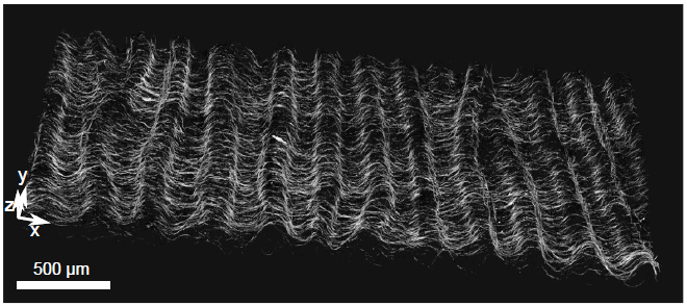
Figure 2: Active matter locally converts chemical energy into mechanical work and, for this reason, it provides new mechanisms of pattern formation. In particular, active nematic fluids made of protein motors and filaments are far-from-equilibrium systems that may exhibit spontaneous motion. We demonstrated that a 3D solution of kinesin motors and microtubule filaments can also spontaneously form a 2D free-standing nematic active sheet that actively buckles out-of-plane into a centimeter-sized periodic corrugated sheet that is stable for days.
Recent publications
- Sarfati G, Maitra A, Voituriez R, Galas J-C, Estévez-Torres A, Crosslinking and depletion determine spatial instabilities in cytoskeletal active matter, Soft Matter, 2022
- Galas J-C, Estévez-Torres A, Van Der Hofstadt M, Long-Lasting and Responsive DNA/Enzyme-Based Programs in Serum-Supplemented Extracellular Media, ACS Synthetic Biology, 2022
- del Junco C, Estévez-Torres A, Maitra A, Front speed and pattern selection of a propagating chemical front in an active fluid, Physical Review E, 2022
- Senoussi A, Galas JC, Estévez-Torres A, Programmed mechano-chemical coupling in reaction-diffusion active matter, Science Advances, 2021
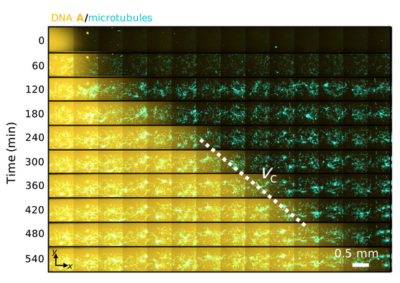
- Vyborna Y, Galas JC, Estévez-Torres A, DNA-Controlled Spatiotemporal Patterning of a Cytoskeletal Active Gel, JACS, 2021
- Senoussi A, Vyborna Y, Berthoumieux H, Galas JC, Estévez-Torres A, Learning from Embryo Development to Engineer Self-organizing Materials chapter in 'Out-of-Equilibrium (Supra)molecular Systems and Materials' book (Editor: Nicolas Giuseppone, Andreas Walther), Wiley, 2021
- Van Der Hofstadt M, Galas J-C, Estévez-Torres A, Spatiotemporal Patterning of Living Cells with Extracellular DNA Programs,
ACS Nano, 2021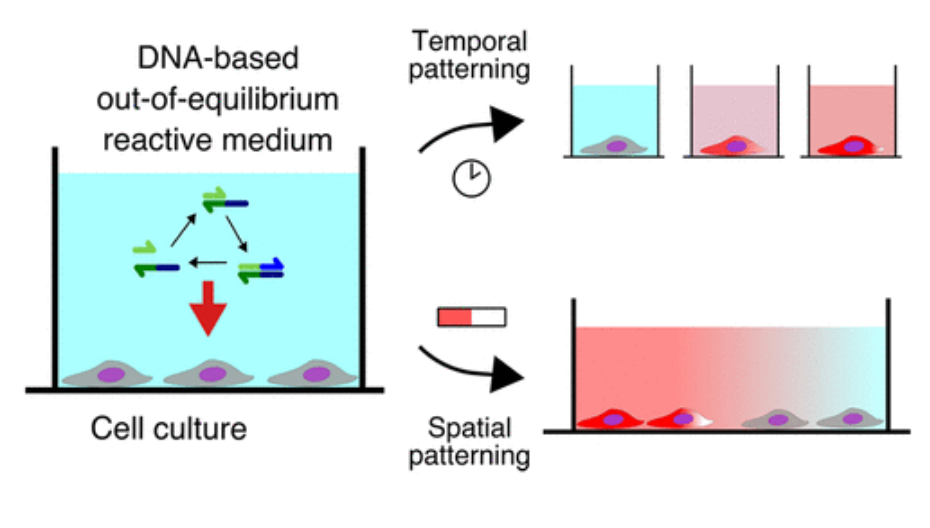
- Senoussi A, Kashida S, Voituriez R, Galas J-C, Maitra A, Estévez-Torres A, Tunable corrugated patterns in an active gel sheet, PNAS, 2019 Preprint on arxiv
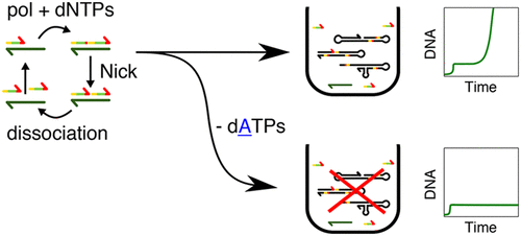
- Urtel G, Van Der Hofstadt M, Galas J-C, Estévez-Torres A,
rEXPAR: an isothermal amplification scheme that is robust to autocatalytic parasites,
ACS Biochemistry, 2019 Preprint on arxiv
- A. Zadorin, Y. Rondelez, G. Gines, V. Dilhas, G. Urtel, A. Zambrano, J.-C. Galas, A. Estevez-Torres, Synthesis and materialization of a reaction-diffusion French flag pattern, Nature chem., 2017, doi:10.1038/nchem.2770. Preprint on arxiv
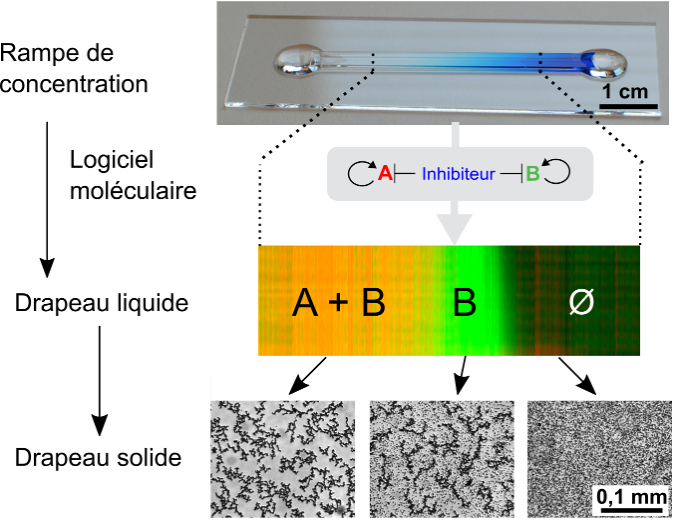
- G. Gines, A. Zadorin, J.-C Galas, T. Fujii, A. Estevez-Torres, Y. Rondelez, Microscopic agents programmed by DNA circuits, Nature nano, 2017, doi:10.1038/nnano.2016.299
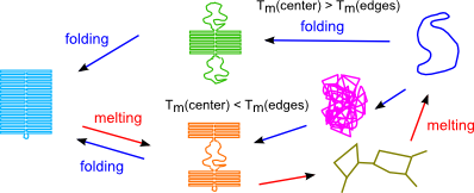
- Lee Tin Wah J, David C, Rudiuk S, Baigl D, Estévez-Torres, A, Observing and Controlling the Folding Pathway of DNA Origami at the Nanoscale, ACS nano, 2016.
- Estevez-Torres A, Biophysics: The expressionist movement, Nature Physics, 2015. News and views.
- Zambrano A, Zadorin AS, Rondelez Y, Estevez-Torres A, Galas JC, Pursuit-and-evasion reaction- diffusion waves in microreactors with tailored geometry, J. Phys. Chem. B, 119(17) :5349-5355, 2015.
- Zadorin AS, Rondelez Y, Galas J-C, & Estevez-Torres A, Synthesis of programmable reaction-diffusion fronts using DNA catalyzers. Phys. Rev. Lett. 2015 114(6). Highlighted in Nature nanotech, Physics and Chemistry world.
You may find further information in our detailed webpage: http://ljpmorpho.free.fr
and you may check our youtube channel