|
Toward microscopic simulations of the cellular environment
Par Fabio Sterpone (IBPC, Laboratoire de Biochimie Théorique, Paris)
Le 8 Décembre 2015 à 10h00 - Salle de séminaires 5ème étage, Tour 32-33
|
Résumé
Proteins work in an extremely heterogeneous and crowded environment. Indeed 5 to 40 % of the intracellular total volume is occupied by large biomolecules [1]. This crowding condition affects both the mobility and stability of a protein. Understanding the molecular details of protein’s life in the cell is therefore considered a challenge for modern biophysics and molecular biology [2]. Unfortunately, the experimental manipulation of the intracellular milieu is very difficult, as well as the in silico characterization of systems with such a large size and spread of length- and time-scales [3].
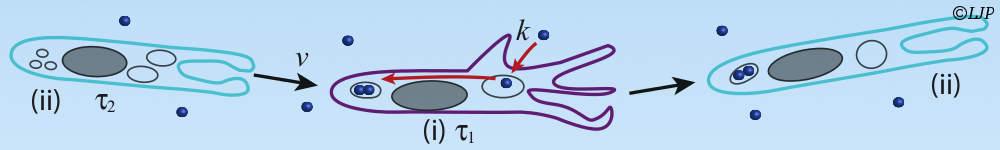
We have recently developed a multi-scale multi-physics approach that enables us to investigate proteins in cell like environments via computer simulations [4]. This approach combines molecular and fluid dynamics and allows to include naturally hydrodynamic interactions in water free coarse-grained model for biomolecules at the quasi-atomistic resolution. In this talk we will describe the methodology and showcase the ensemble of applications currently on the way. This includes the study of amyloid peptide aggregation (the hallmark of neurodegenerative diseases like Alzheimer), the protein unfolding under shear flow, the mobility of proteins in crowded environment.
[1] R. J. Ellis, A. P. Minton, Nature 2003, 425,27-28.
[2] a) I.Guzman,M.Gruebele, J Phys Chem 2014,118,8459. b) M.Sarkar,A.E. Smith, G.J. PNAS 2013, 110, 19342-19347.
[3] S.R. McGuffee, A. H. Elcock, PLoS Comput Biol 2010, 6,e1000694.
[4] a) F.Sterpone, P. Derreumaux, S. Melchionna, J Chem Theory Comput 2015, 11, 1843-1853. b) F. Sterpone et al, Chem Soc Rev 2014, 43, 4871-4893.