|
High-resolution mapping of bifurcations in nonlinear biochemical circuits
Par Anthony Genot (LIMMS/U. Tokyo, Japan)
Le 10 Avril 2018 à 11h00 - Salle de séminaires 5ème étage, Tour 32-33
|
Résumé
DNA has emerged in the past decades as a versatile and powerful polymer to build and program at the nanoscale. A rich library of devices has been built with DNA: origamis [1], biochemical neural networks [2], or reconfigurable plasmonic structures [3]. This DNA nanotechnology paves the route to societal applications such as smart therapeutics, autonomous chemical assembly lines or point-of-care diagnosis.
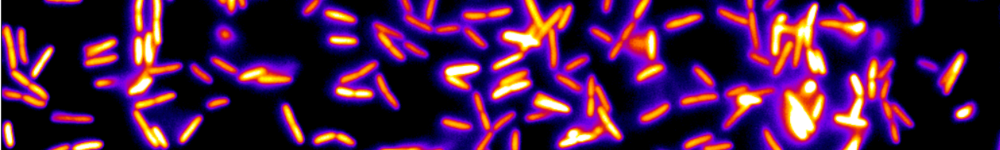
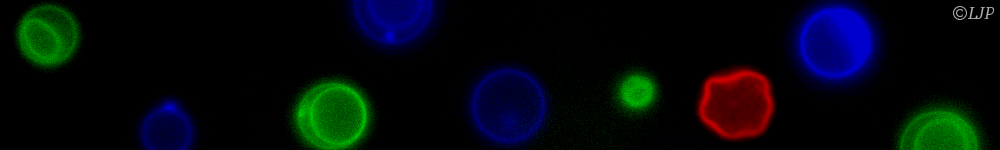
In LIMMS (CNRS/University of Tokyo), we recently developed a DNA-based toolbox to implement complex chemical dynamics, such as bistability or oscillations, with DNA and 3 enzymes [4]. But those nonlinear systems are extremely sensitive to their design parameters (concentrations of enzymes and DNA), which complicates design and characterisation. To tackle the resulting combinatorial explosion, we designed a droplet-based microfluidic platform to prepare tens of thousands of droplets with various combinations of parameters, allowing us to scan the parameter space efficiently [5].
Compared to conventional biochemical tools, the platform increased experimental throughput by 2 orders of magnitudes (from 100 to 10,000 data points/day) and reduced volumes by 5 orders of magnitudes (from 10 μL to 100 pL per data point).
We used this unprecedented resolution to finely study the dynamic of the bistable switch and a DNA oscillator, revealing new insights about the biochemical mechanisms at play.
[1] Rothemund, Nature 440.7082 (2006): 297-302.
[2] Qian et al., Nature 475.7356 (2011): 368-372.
[3] Kuzyk, Anton, et al., Nature materials 13.9 (2014): 862-866.
[4] Padirac et al., P.N.A.S. 109.47 (2012): E3212-E3220.
[5] Genot, A. J., et al., Nature Chemistry (2016).