|
Programmable self-assembly of nanotubes using DNA origami colloids
Par Daichi Hayakawa - Rogers Lab, Brandeis University
Le 23 Avril 2024 à 11h00 - Laboratoire Jean Perrin - Campus Jussieu - T 32-33 - 5e et. - P533
|
Résumé
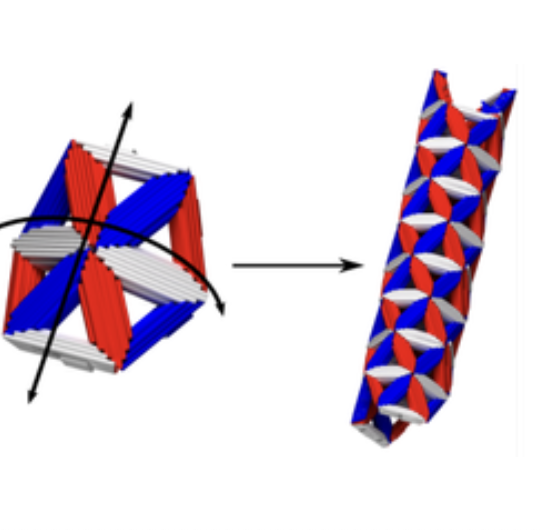
Nature is replete with self-assembled materials that have one or more self-limited dimensions, including shells, tubules, and fibers. Despite significant advances in making nanometer- and micrometer-scale subunits, the programmable assembly of similar self-limiting architectures from synthetic components has remained largely out of reach, mainly due to difficulty in tuning the valence, binding angle, and interaction specificity, simultaneously. In this talk, I will show how we create geometrically programmed subunits using DNA origami and study their assembly into tubules with a self-limited width. We show that the width and the helicity of tubules can be tuned by changing the local curvature encoded in a single subunit. Exploiting the programmability of our system, we further test the tradeoffs between two paradigms of self-limited assembly: self-closure through programmed curvature and addressable assembly through programmed, specific interactions. By increasing the number of unique subunits in the system, the allowed combination of width and helicities of the tubules decreases, leading to more specific tubule geometries. In an extreme case, we use 16 unique subunits to assemble a tubule, leading to almost a full selectivity of the tubule geometry. In this talk, I will also describe an inverse design method for assembling periodic tilings, which we used to generate the interaction matrix necessary to assemble the multicomponent tubules.