SEP 2018
André Estevez-Torres a le plaisir de vous inviter à sa soutenance d'habilitation à diriger des recherches qui se tiendra le mardi 4 septembre à 14h00 à la salle de réunion de l'INSP (campus de Jussieu, tour 23, 3eme étage, couloir 23-22, salle 317 - détails d'accès ci-dessous), où il présentera ses travaux sur:
Auto-organisation et auto-assemblage dans des systèmes chimiques à base d'acides nucléiques
La soutenance aura lieu en anglais devant un jury composé de:
-Prof. Roy BAR-ZIV, Weizmann Institute of Science, Rapporteur
-Dr. Nadine PEYRIERAS, CNRS, Rapporteure
-Dr. Benoît LIMOGES, CNRS and Université Paris Diderot, Rapporteur
-Dr. Cecile SYKES, Institut Curie, CNRS and Sorbonne Université, Examinatrice
-Prof. Thomas SURREY, The Francis Crick Institute, Examinateur
-Prof. Anne DE WIT, Université libre de Bruxelles, Examinatrice
Abstract
The question that motivates my research is to what extent molecular systems can create spatial order at the macroscopic scale. In this context, two types of mechanisms should be considered: self-organization, that dissipates energy, and self-assembly, that does not. Self-assembly is mainly controlled by thermodynamics while self-organization is ruled by kinetics. Because both the thermodynamics and the kinetics of chemical reactions are difficult to predict, I have naturally investigated these processes using a unique class of molecules for which both can be predicted from the structure: nucleic acids (DNA and RNA).
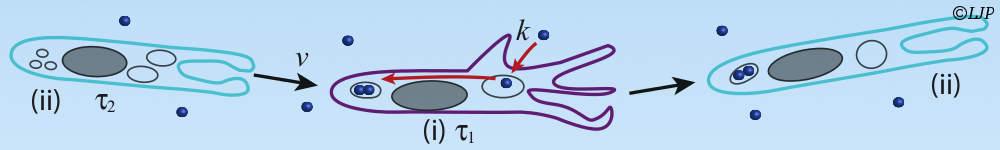
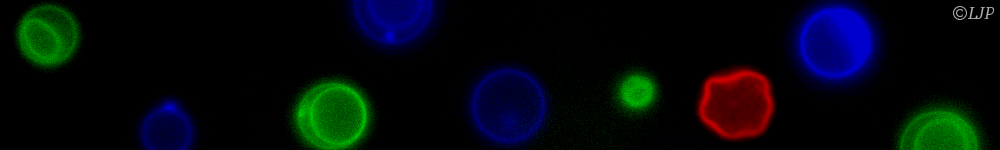
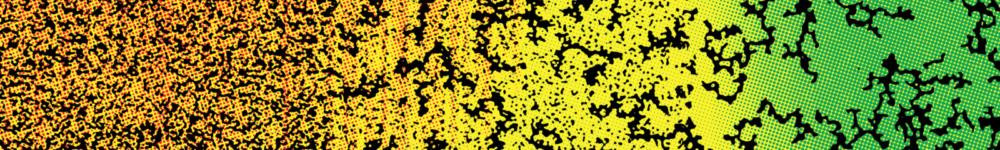
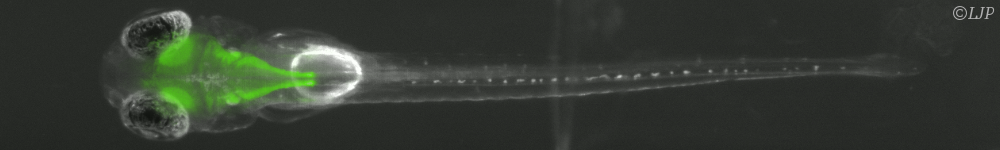
Concerning self-organization, our efforts have focused on the experimental design of reaction-diffusion patterns. To do so we have used a set of highly reconfigurable synthetic chemical reaction networks based on DNA that can be kept out of equilibrium for days. I will first describe the advantages of this set of reactions to synthesize different spatio-temporal patterns, such as travelling waves and stationary fronts. I will then show how these patterns can be exploited for devising synthetic materials inspired from embryonic development. I will end this part by suggesting open questions in physics and chemistry that could be addressed in a near future using this programmable biochemical system.
In addition, we have developed methods to characterize important self-assembly processes. On the one hand, we used AFM to investigate the folding pathway of DNA origami. On the other hand, we employed cell-free transcription-translation systems to characterize the thermodynamics of synthetic riboregulators.