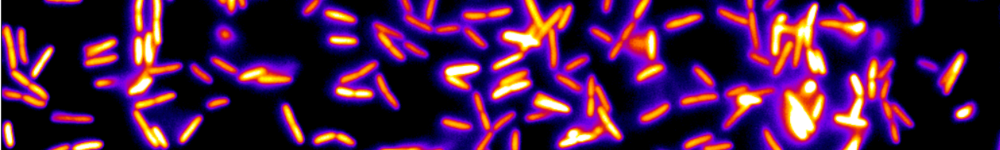
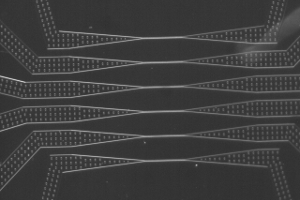
Le LJP en bref
L'ensemble des activités du LJP est tourné vers la physique à l'interface avec la biologie et la médecine. Nous dévelopons de nouvelles approches expérimentales visant à sonder les propriétés de systèmes biologiques complexes à différentes échelles, et nous cherchons à réaliser de nouveaux systèmes physico-chimiques dont les comportements pourraient offrir certaines analogies avec des systèmes biologiques. Une grande partie de nos recherches se situe dans le contexte de la réponse d’un système biologique à des perturbations extérieures. Le laboratoire comprend aussi une composante théorique en constante interaction avec nos activités expérimentales.
Prochain séminaire
Tous les séminaires|
30
AVR 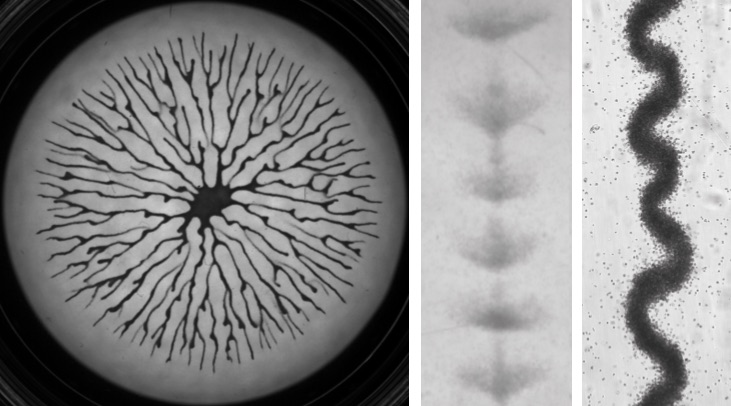
|
Shedding light on micro-algae: phototaxis-induced collective phenomena
Par Raphaël Jeanneret - LPENS, Physics Department ENS
Le 30 Avril 2024 à 11h00 - Laboratoire Jean Perrin - Campus Jussieu - T 32-33 - 5e et. - P533
Motile micro-algae modify their environment by absorbing light, consuming and releasing chemical compounds and generating flows. Flows, light, and chemicals in turn influence their motion. These ...
|
Zoom sur ...
Actualités
Toutes les actualités
26
JAN 2024
JAN 2024
Soutenance de thèse de SEOW Vui Yin (Mafer)
Je vous invite à ma soutenance de thèse sur "Transformation de l'ADN et pili de type IV ...
Je vous invite à ma soutenance de thèse sur "Transformation de l'ADN et pili de type IV ...

20
JUN 2023
JUN 2023
Soutenance de thèse de Guillaume Sarfati
J'ai le plaisir de vous inviter à ma soutenance de thèse qui aura lieu le mardi 20 juin ...
J'ai le plaisir de vous inviter à ma soutenance de thèse qui aura lieu le mardi 20 juin ...

13
JUN 2023
JUN 2023
Retaite du LJP (édition 2023)
La retraite du LJP a eu lieu du 13 au 15 juin 2023 à la Chancellerie de Montrichard (Loire-et-Cher). ...
La retraite du LJP a eu lieu du 13 au 15 juin 2023 à la Chancellerie de Montrichard (Loire-et-Cher). ...

06
AVR 2023
AVR 2023
Soutenance de thèse d'Hippolyte Moulle - 06/04/2023 à 9h30 - IBPS salle B501
I am glad to invite you to my PhD defense, which will take place on Thursday the 6th of April at 9:30AM, in ...
I am glad to invite you to my PhD defense, which will take place on Thursday the 6th of April at 9:30AM, in ...
09
MAR 2023
MAR 2023
Soutenance de thèse de Nicolas Escoubet - 09/03/2023 à 14h00 - Amphithéâtre ...
Nicolas Escoubet soutiendra sa thèse Jeudi 09 Mars 2023 à 14h00 dans l'amphithéâtre...
Nicolas Escoubet soutiendra sa thèse Jeudi 09 Mars 2023 à 14h00 dans l'amphithéâtre...

08
DÉC 2022
DÉC 2022
Décès de Jean Cognet
C’est avec une immense tristesse que nous avons appris le décès de Jean Cognet, ...
C’est avec une immense tristesse que nous avons appris le décès de Jean Cognet, ...
Dernières publications
Toutes les publications| ⊞ | DiSTNet2D: Leveraging Long-Range Temporal Information for Efficient Segmentation and Tracking - PRX Life (Apr. 2024) |
| ⊞ | Identification of extracellular vesicles from their Raman spectra via self-supervised learning - Scientific Reports (Mar. 2024) |
| ⊞ | Structure and individuality of navigation in zebrafish larvae - hal (Feb. 2024) |
| ⊞ | Lipidomics and biodistribution of extracellular vesicles-secreted by hepatocytes from Zucker lean and fatty rats - Journal of Extracellular Biology (Feb. 2024) |
| ⊞ | Extracellular vesicles from activated platelets possess a phospholipid-rich biomolecular profile and enhance prothrombinase activity - Journal of Thrombosis and Haemostasis (Jan. 2024) |
|
|
|
Bibtex | doi:10.1016/j.jtha.2024.01.004 |