Biophysique des micro-organismes
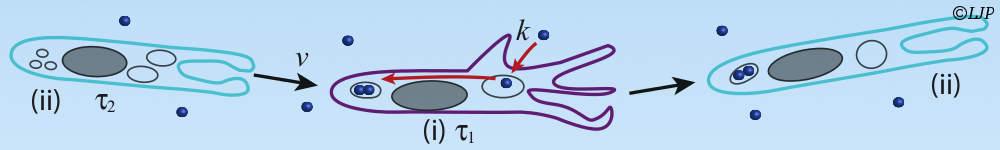
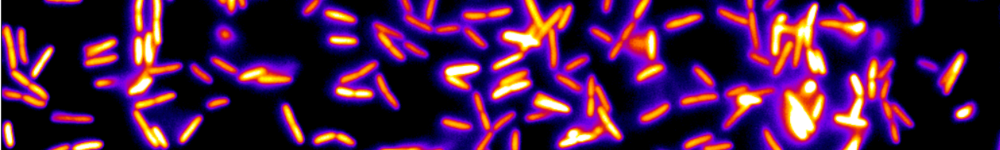
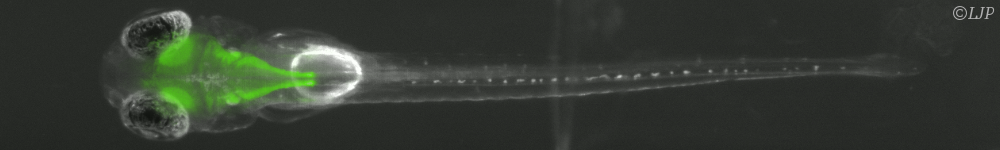
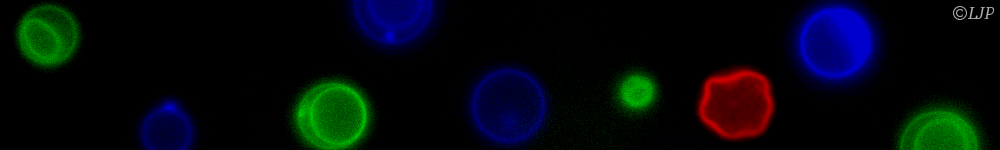
Les micro-organismes constituent l’essentiel de la biomasse terrestre et présentent une immense diversité de formes, de comportements et de biotopes. Nous nous intéressons plus particulièrement aux bactéries. Leur temps de génération (de l’ordre de l’heure) et leur taille (de l’ordre du micromètre) permettent de les explorer de l’échelle de la cellule unique à celle des populations composites organisées en société. Nous étudions ces systèmes sur la base du développement de dispositifs microfluidiques microfabriqués dédiés, couplé aux avancées les plus récentes de la génétique et à des approches de vidéo-microscopie et d’imagerie de fluorescence. Notre objectif est de comprendre les mécanismes qui sous-tendent les comportements de ces systèmes dans des environnements contrôlés en étudiant les relations causales de leurs propriétés physiques, physico-chimiques et biologiques.






Doctorants (soutenance 2018->)
-
2025 Martin Maliet
-
2023 Vui-Yin Seow
-
2022 Wafa Ben Youssef
-
2019 Amaury Monmeyran
-
2018 Alexandre Deloupy
Post-docs
- 2023- Gaspard Junot
- 2020-2021 Jerko Rosko
- 2015-2019 Jean Ollion
Stagiaires Master
Master 1:
- 2020 Valentin Bonnet
- 2021 Assia Benachir
- 2022 Elif Uyar
- 2023 Jinghan Huang
- 2023 Adrien Bérard
Master 2:
- 2018 Yvan Bourrigault
- 2018 Wafa Benyoussef
- 2019 Raphaël Monteil
- 2021 Candice hermant
- 2020 Elif Elçin
- 2022 Nicolas Pellet
- 2022 Martin Maliet
- 2022 Naicen Naourra
Autre page reliée
Cycle cellulaireOffres d'emploi associées
2024
2024
2024
Publications
2024
| ⊞ | DiSTNet2D: Leveraging Long-Range Temporal Information for Efficient Segmentation and Tracking - PRX Life (Apr. 2024) |
2023
| ⊞ | Direct comparison of spatial transcriptional heterogeneity across diverse Bacillus subtilis biofilm communities - Nature communications (Nov. 2023) |
| ⊞ | DistNet2D: Leveraging long-range temporal information for efficient segmentation and tracking - Arxiv (Oct. 2023) |
| ⊞ | Multisite transformation in Neisseria gonorrhoeae: insights on transformations mechanisms and new genetic modification protocols - Frontiers in Microbiology (Jun. 2023) |
| ⊞ | Emergence of Exopolysaccharides Overproducers Is Linked to Environmental Spatial Structure via Redox State - mSphere (Apr. 2023) |
| ⊞ | Long-range alteration of the physical environment mediates cooperation between Pseudomonas aeruginosa swarming colonies - Environmental Microbiology (Mar. 2023) |
2022
| ⊞ | Flagellar Motility During E. coli Biofilm Formation Provides a Competitive Disadvantage Which Recedes in the Presence of Co-Colonizers - Front Cell Infect Microbiol (Jul. 2022) |
| URL | Full text PDF | Bibtex | doi:10.3389/fcimb.2022.896898 |
| ⊞ | Spatial-temporal dynamics of a microbial cooperative behavior resistant to cheating - Nature communications (Jun. 2022) |
|
|
|
Bibtex | doi:https://doi.org/10.1038/s41467-022-28321-9 |
2021
| ⊞ | An In Vitro Model System to Test Mechano-Microbiological Interactions Between Bacteria and Host Cells - Cytoskeleton : Methods and Protocols (Sep. 2021) |
| ⊞ | Cooperation between melanoma cell states promotes metastasis through heterotypic cluster formation - Developmental Cell (Aug. 2021) |
| ⊞ | Fresh Extension of Vibrio cholerae Competence Type IV Pili Predisposes Them for Motor-Independent Retraction - Applied and Environmental Microbiology (Jun. 2021) |
| ⊞ | Open science saves lives: lessons from the COVID-19 pandemic - BMC MEDICAL RESEARCH METHODOLOGY (Jun. 2021) |
| ⊞ | Four species of bacteria deterministically assemble to form a stable biofilm in a millifluidic channel - NPJ Biofilms Microbiomes (Jan. 2021) |
| URL | Full text PDF | Bibtex | doi:10.1038/s41522-021-00233-4 |
2020
| ⊞ | Spatiotemporal pattern formation in E. coli biofilms explained by a simple physical energy balance - Soft Matter (Jan. 2020) |
| URL | Full text PDF | Bibtex | doi:10.1039/c9sm01375j |
2019
| ⊞ | Evolution at the edge of expanding populations - The American Naturalist (Sep. 2019) |
2018
| ⊞ | Environmentally Mediated Social Dilemmas - Trends in Ecology and Evolution (Nov. 2018) |
| ⊞ | The inducible chemical-genetic fluorescent marker FAST outperforms classical fluorescent proteins in the quantitative reporting of bacterial biofilm dynamics - Scientific Reports (Oct. 2018) |
| URL | Full text PDF | Bibtex | doi:10.1038/s41598-018-28643-z |
| ⊞ | Intermittent Pili-Mediated Forces Fluidize Neisseria meningitidis Aggregates Promoting Vascular Colonization - Cell (Jun. 2018) |
| ⊞ | Archaeal cells share common size control with bacteria despite noisier growth and division - Nature Microbiology (Feb. 2018) |
|
|
|
Bibtex | doi:10.1038/s41564-017-0082-6 |
| ⊞ | Method for Detecting and Studying Genome-Wide Mutations in Single Living Cells in Real Time - Methods Mol Biol (Jan. 2018) |
|
|
|
Bibtex | doi:https://doi.org/10.1007/978-1-4939-7638-6_3 |
2017
| ⊞ | Bacterial biofilm under flow: First a physical struggle to stay, then a matter of breathing - PLOS ONE (Apr. 2017) |
| URL | Full text PDF | Bibtex | doi:10.1371/journal.pone.0175197 |
2016
| ⊞ | Single-cell analysis of growth in budding yeast and bacteria reveals a common size regulation strategy - Current Biology (Feb. 2016) |
|
|
|
Bibtex | doi:10.1016/j.cub.2015.11.067 |
2015
| ⊞ | Single-Cell Analysis of Growth and Cell Division of the Anaerobe Desulfovibrio vulgaris Hildenborough - Frontiers in Microbiology (Dec. 2015) |
|
|
|
Bibtex | doi:10.3389/fmicb.2015.01378 |
| ⊞ | Size sensors in bacteria, cell cycle control, and size control - Frontiers in Microbiology (May. 2015) |
|
|
|
Bibtex | doi:10.3389/fmicb.2015.00515 |
| ⊞ | Statistical estimation of a growth-fragmentation model observed on a genealogical tree - Bernoulli (Jan. 2015) |
|
|
|
Bibtex | doi:10.3150/14-BEJ623 |
2014
| ⊞ | Division in Escherichia coli is triggered by a size-sensing rather than a timing mechanism - BMC Biology (Feb. 2014) |
|
|
|
Bibtex | doi:10.1186/1741-7007-12-17 |